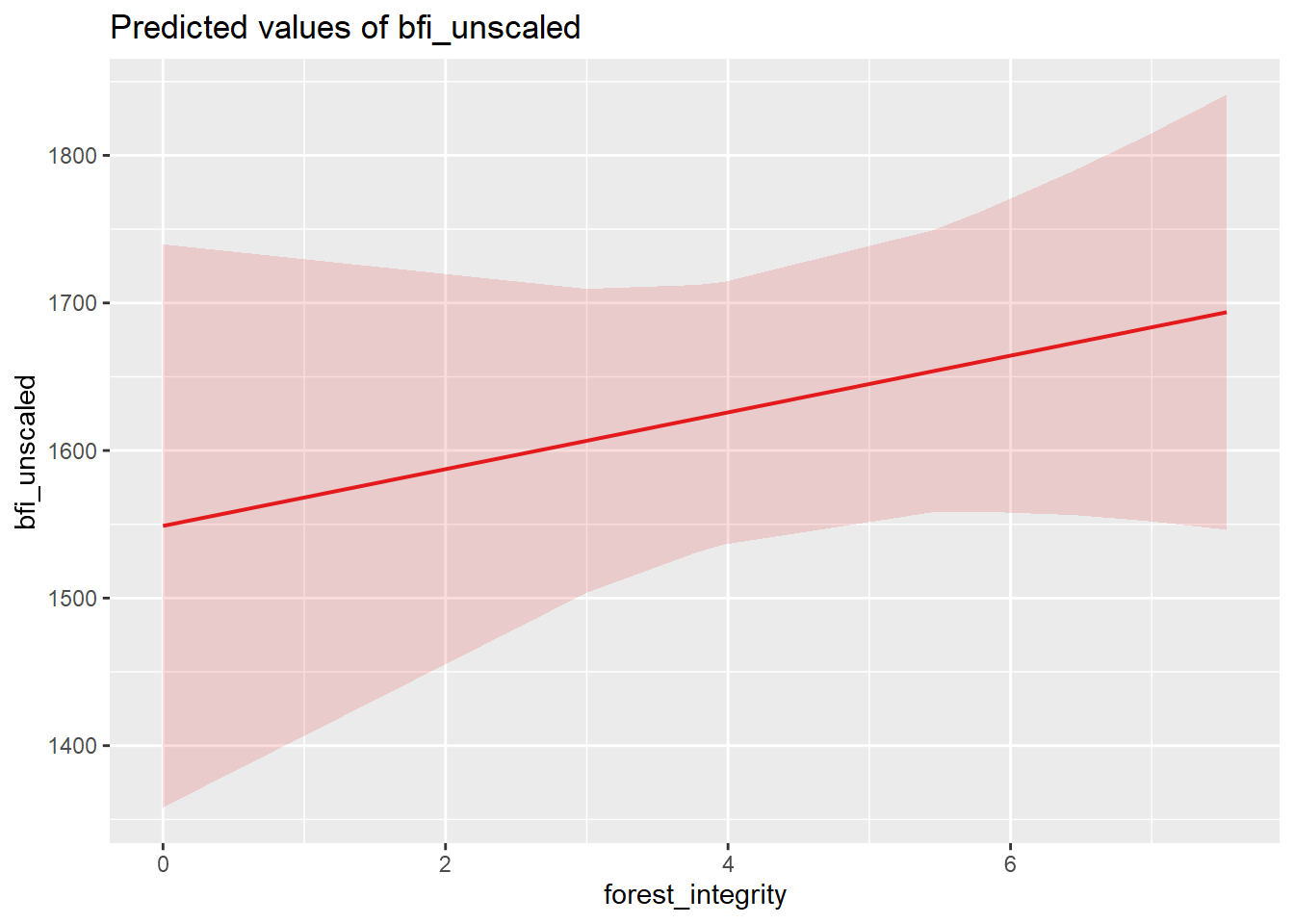
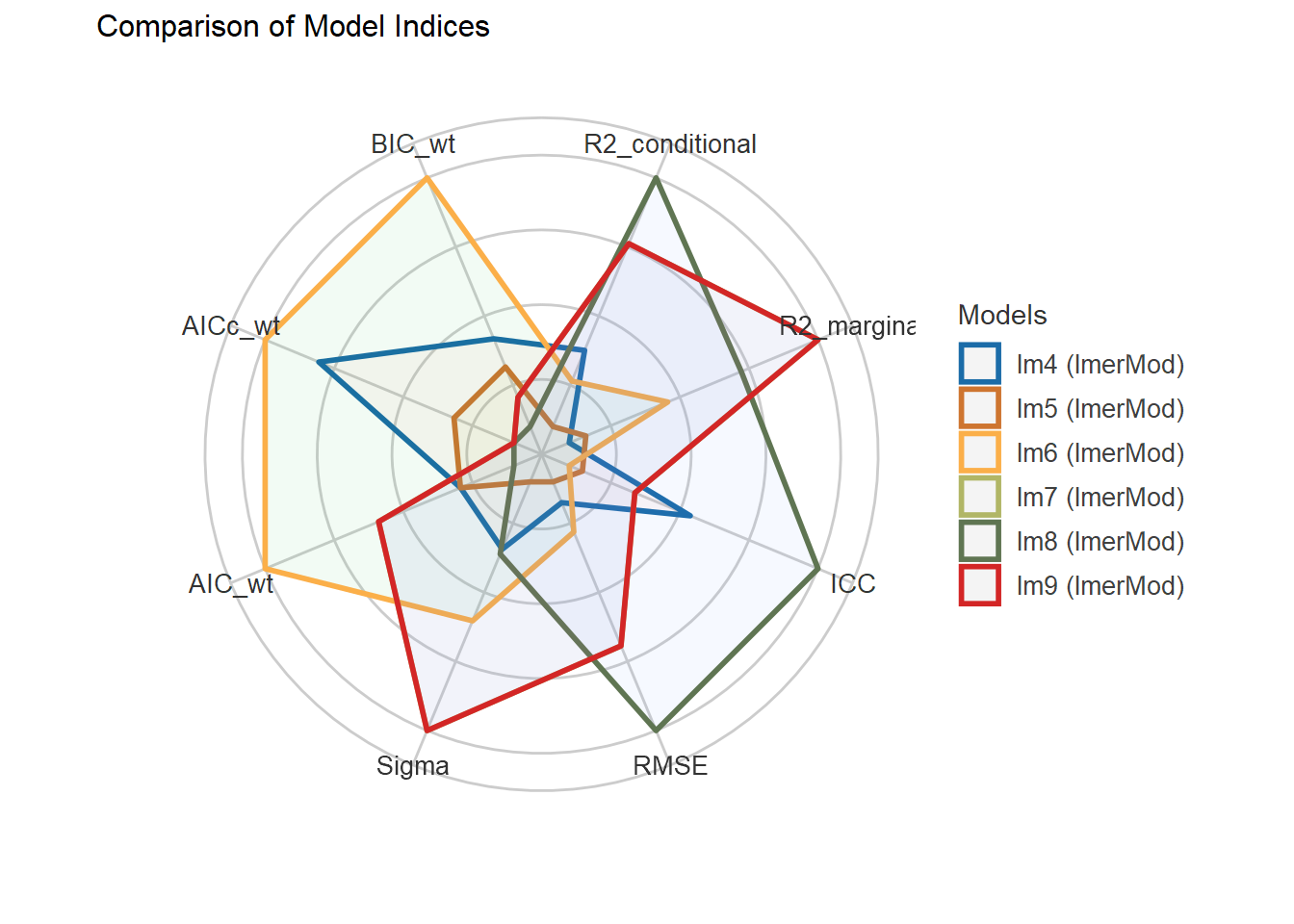
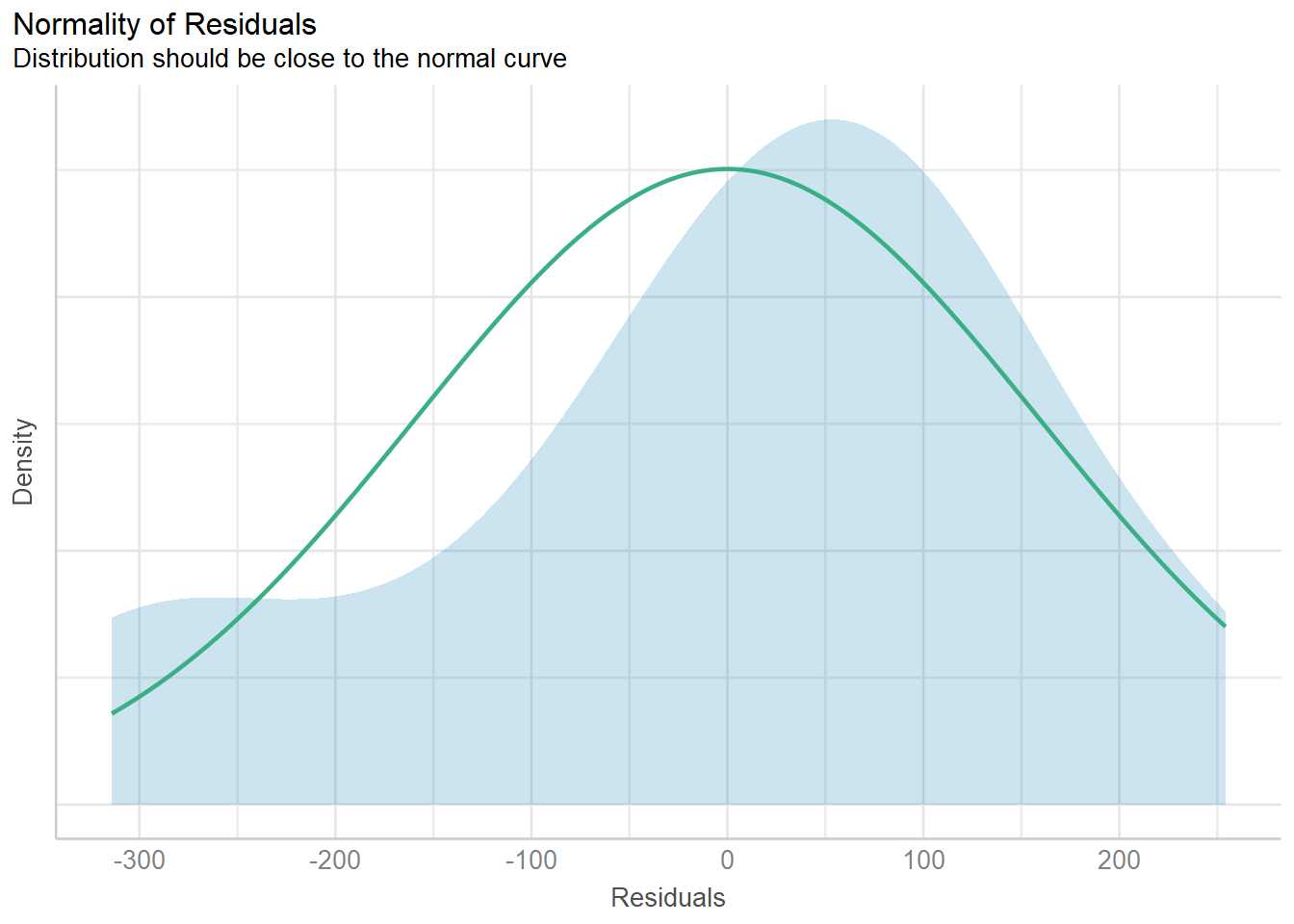
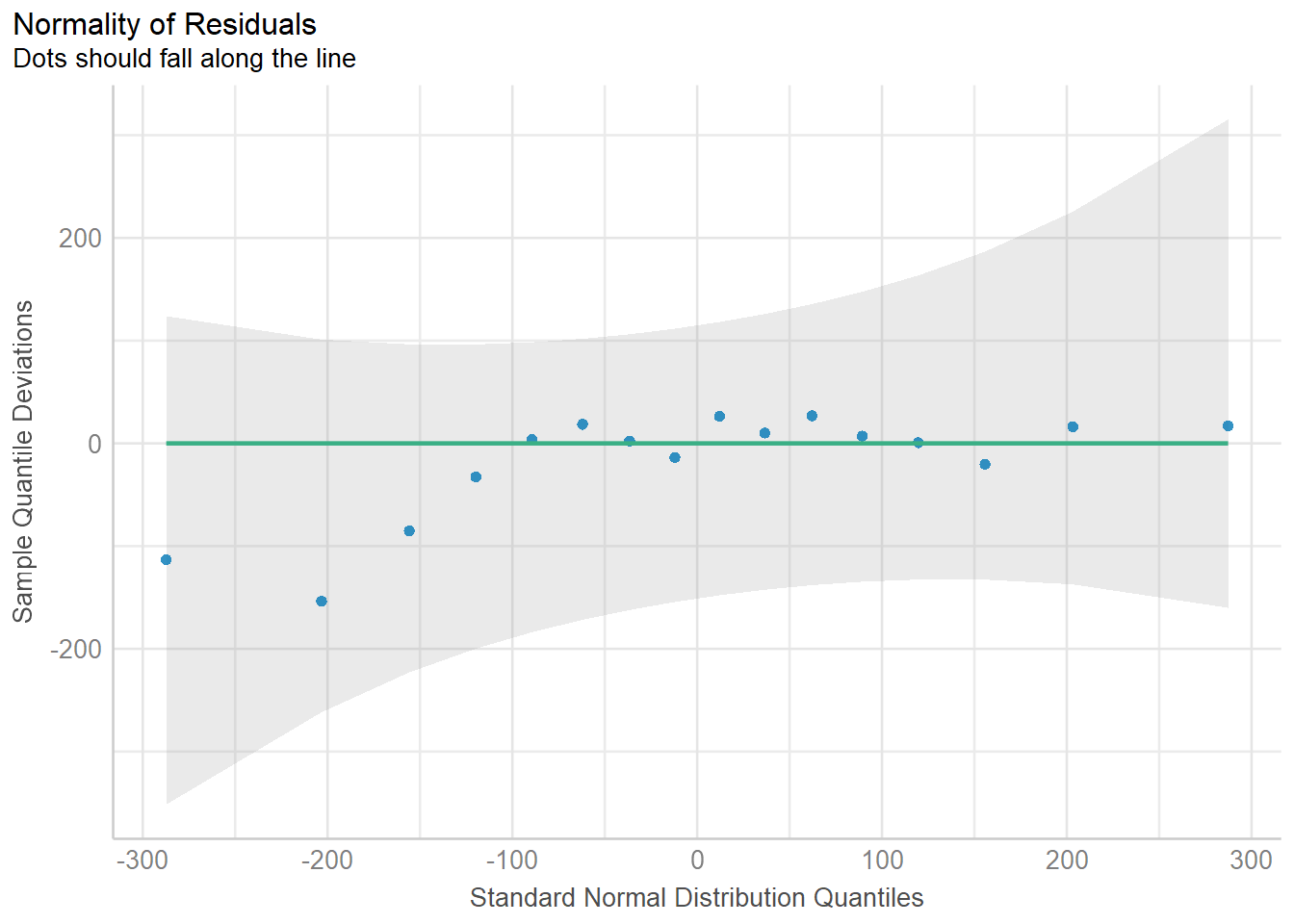
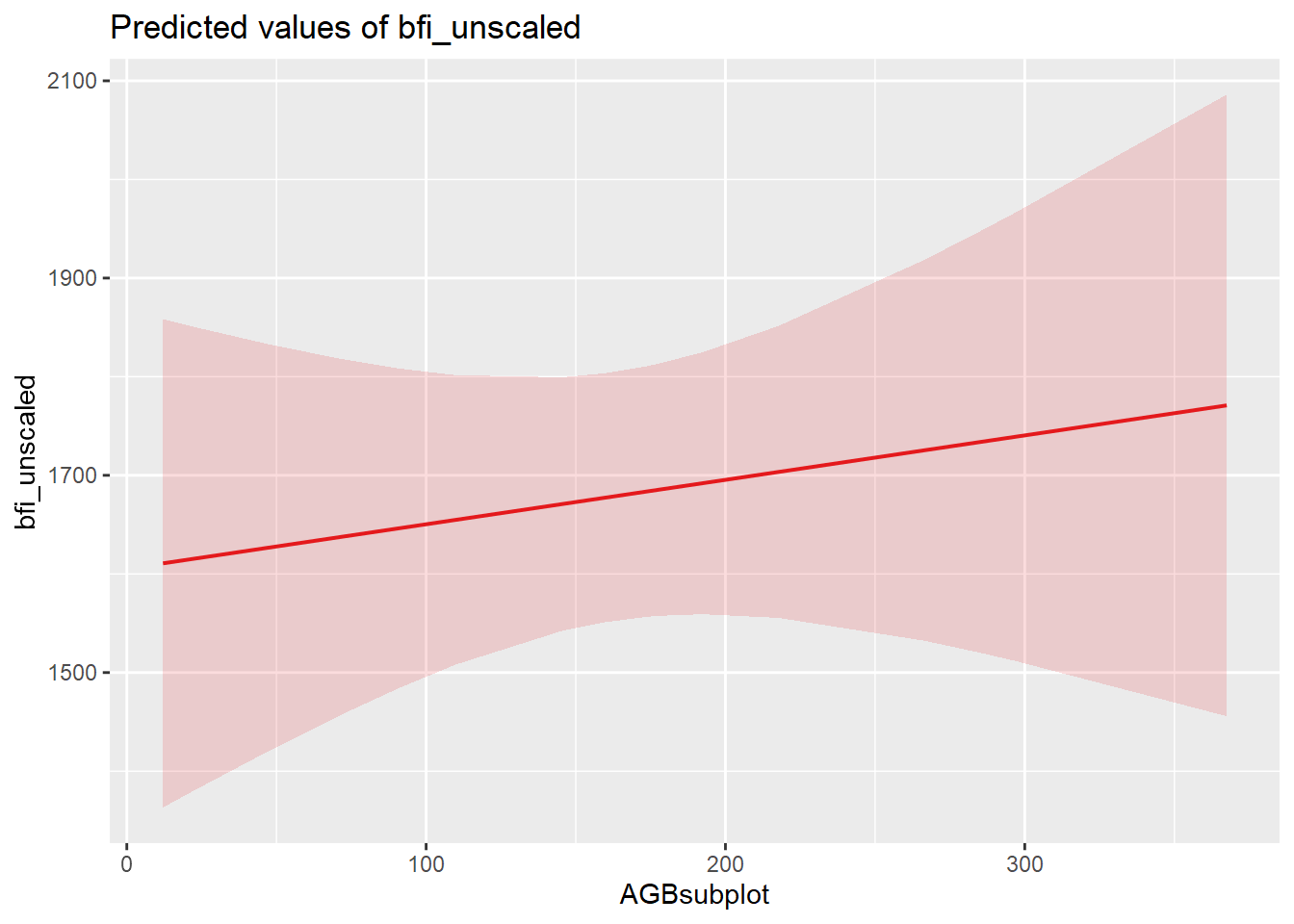
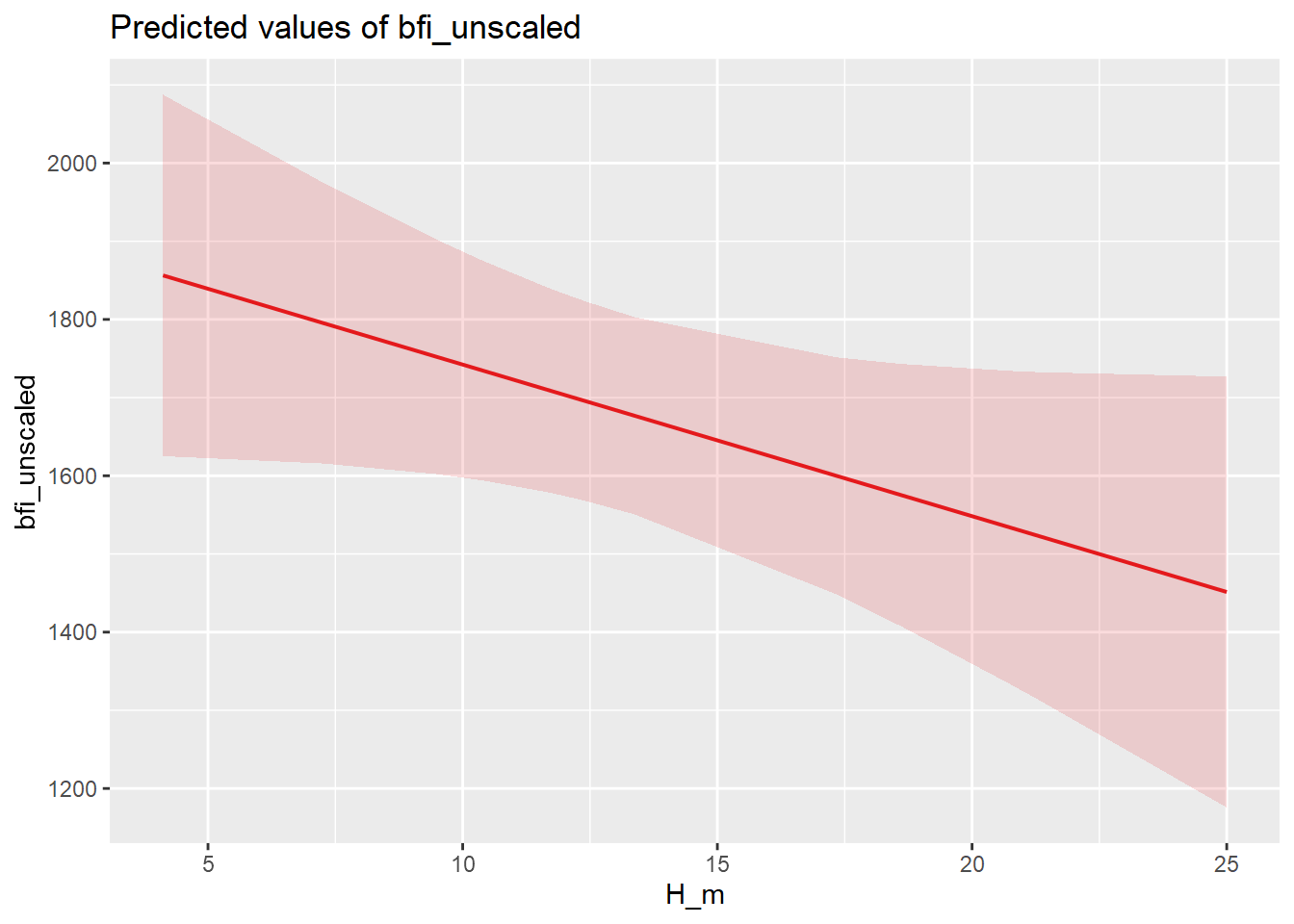
| Bartoń K (2023). MuMIn: Multi-Model Inference. R package version 1.47.5, https://CRAN.R-project.org/package=MuMIn. |
| Bates D, Mächler M, Bolker B, Walker S (2015). “Fitting Linear Mixed-Effects Models Using lme4.” Journal of Statistical Software, 67(1), 1-48. doi:10.18637/jss.v067.i01 https://doi.org/10.18637/jss.v067.i01. |
| Bates D, Maechler M, Jagan M (2023). Matrix: Sparse and Dense Matrix Classes and Methods. R package version 1.6-1.1, https://CRAN.R-project.org/package=Matrix. |
| Breheny P, Burchett W (2017). “Visualization of Regression Models Using visreg.” The R Journal, 9(2), 56-71. |
| Francisco Rodriguez-Sanchez, Connor P. Jackson (2023). grateful: Facilitate citation of R packages. https://pakillo.github.io/grateful/. |
| Gilbert P, Varadhan R (2019). numDeriv: Accurate Numerical Derivatives. R package version 2016.8-1.1, https://CRAN.R-project.org/package=numDeriv. |
| Grolemund G, Wickham H (2011). “Dates and Times Made Easy with lubridate.” Journal of Statistical Software, 40(3), 1-25. https://www.jstatsoft.org/v40/i03/. |
| Hijmans R (2024). terra: Spatial Data Analysis. R package version 1.7-71, https://CRAN.R-project.org/package=terra. |
| Hijmans RJ, Barbosa M, Ghosh A, Mandel A (2023). geodata: Download Geographic Data. R package version 0.5-9, https://CRAN.R-project.org/package=geodata. |
| Iannone R, Cheng J, Schloerke B, Hughes E, Lauer A, Seo J (2024). gt: Easily Create Presentation-Ready Display Tables. R package version 0.10.1, https://CRAN.R-project.org/package=gt. |
| Lüdecke D (2018). “ggeffects: Tidy Data Frames of Marginal Effects from Regression Models.” Journal of Open Source Software, 3(26), 772. doi:10.21105/joss.00772 https://doi.org/10.21105/joss.00772. |
| Lüdecke D (2024). sjPlot: Data Visualization for Statistics in Social Science. R package version 2.8.16, https://CRAN.R-project.org/package=sjPlot. |
| Lüdecke D, Ben-Shachar M, Patil I, Waggoner P, Makowski D (2021). “performance: An R Package for Assessment, Comparison and Testing of Statistical Models.” Journal of Open Source Software, 6(60), 3139. doi:10.21105/joss.03139 https://doi.org/10.21105/joss.03139. |
| Lüdecke D, Patil I, Ben-Shachar M, Wiernik B, Waggoner P, Makowski D (2021). “see: An R Package for Visualizing Statistical Models.” Journal of Open Source Software, 6(64), 3393. doi:10.21105/joss.03393 https://doi.org/10.21105/joss.03393. |
| Makowski D, Lüdecke D, Patil I, Thériault R, Ben-Shachar M, Wiernik B (2023). “Automated Results Reporting as a Practical Tool to Improve Reproducibility and Methodological Best Practices Adoption.” CRAN. https://easystats.github.io/report/. |
| Müller K, Wickham H (2023). tibble: Simple Data Frames. R package version 3.2.1, https://CRAN.R-project.org/package=tibble. |
| Pebesma E, Bivand R (2023). Spatial Data Science: With applications in R. Chapman and Hall/CRC. doi:10.1201/9780429459016 https://doi.org/10.1201/9780429459016, https://r-spatial.org/book/. Pebesma E (2018). “Simple Features for R: Standardized Support for Spatial Vector Data.” The R Journal, 10(1), 439-446. doi:10.32614/RJ-2018-009 https://doi.org/10.32614/RJ-2018-009, https://doi.org/10.32614/RJ-2018-009. |
| R Core Team (2023). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. |
| Venables WN, Ripley BD (2002). Modern Applied Statistics with S, Fourth edition. Springer, New York. ISBN 0-387-95457-0, https://www.stats.ox.ac.uk/pub/MASS4/. |
| Viechtbauer W (2010). “Conducting meta-analyses in R with the metafor package.” Journal of Statistical Software, 36(3), 1-48. doi:10.18637/jss.v036.i03 https://doi.org/10.18637/jss.v036.i03. |
| Wei T, Simko V (2021). R package ‘corrplot’: Visualization of a Correlation Matrix. (Version 0.92), https://github.com/taiyun/corrplot. |
| White T, Noble D, Senior A, Hamilton W, Viechtbauer W (2022). metadat: Meta-Analysis Datasets. R package version 1.2-0, https://CRAN.R-project.org/package=metadat. |
| Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. ISBN 978-3-319-24277-4, https://ggplot2.tidyverse.org. |
| Wickham H (2023). forcats: Tools for Working with Categorical Variables (Factors). R package version 1.0.0, https://CRAN.R-project.org/package=forcats. |
| Wickham H (2023). stringr: Simple, Consistent Wrappers for Common String Operations. R package version 1.5.1, https://CRAN.R-project.org/package=stringr. |
| Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H (2019). “Welcome to the tidyverse.” Journal of Open Source Software, 4(43), 1686. doi:10.21105/joss.01686 https://doi.org/10.21105/joss.01686. |
| Wickham H, Bryan J (2023). readxl: Read Excel Files. R package version 1.4.3, https://CRAN.R-project.org/package=readxl. |
| Wickham H, François R, Henry L, Müller K, Vaughan D (2023). dplyr: A Grammar of Data Manipulation. R package version 1.1.4, https://CRAN.R-project.org/package=dplyr. |
| Wickham H, Henry L (2023). purrr: Functional Programming Tools. R package version 1.0.2, https://CRAN.R-project.org/package=purrr. |
| Wickham H, Hester J, Bryan J (2024). readr: Read Rectangular Text Data. R package version 2.1.5, https://CRAN.R-project.org/package=readr. |
| Wickham H, Vaughan D, Girlich M (2024). tidyr: Tidy Messy Data. R package version 1.3.1, https://CRAN.R-project.org/package=tidyr. |
| Xie Y, Cheng J, Tan X (2024). DT: A Wrapper of the JavaScript Library ‘DataTables’. R package version 0.32, https://CRAN.R-project.org/package=DT. |