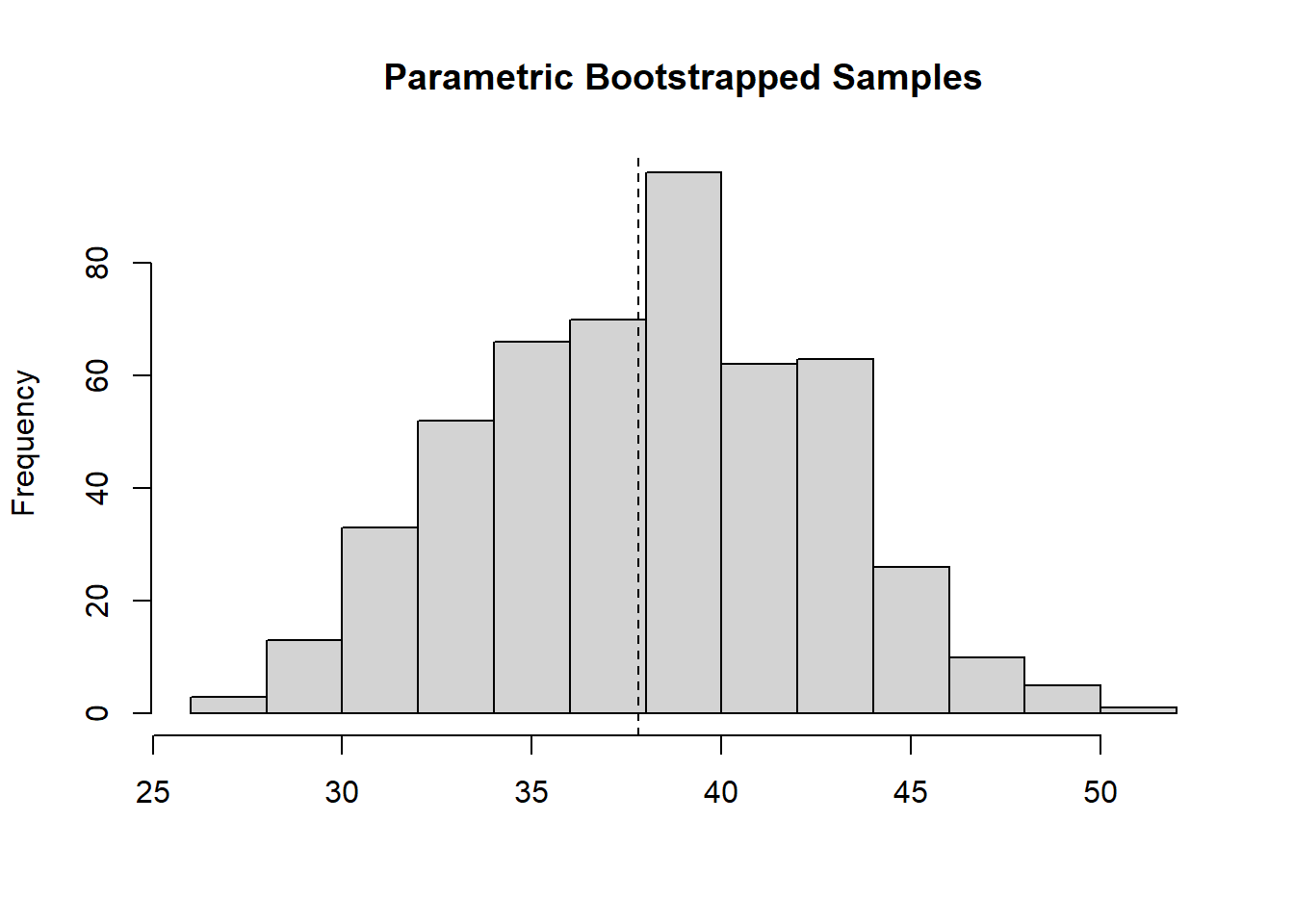
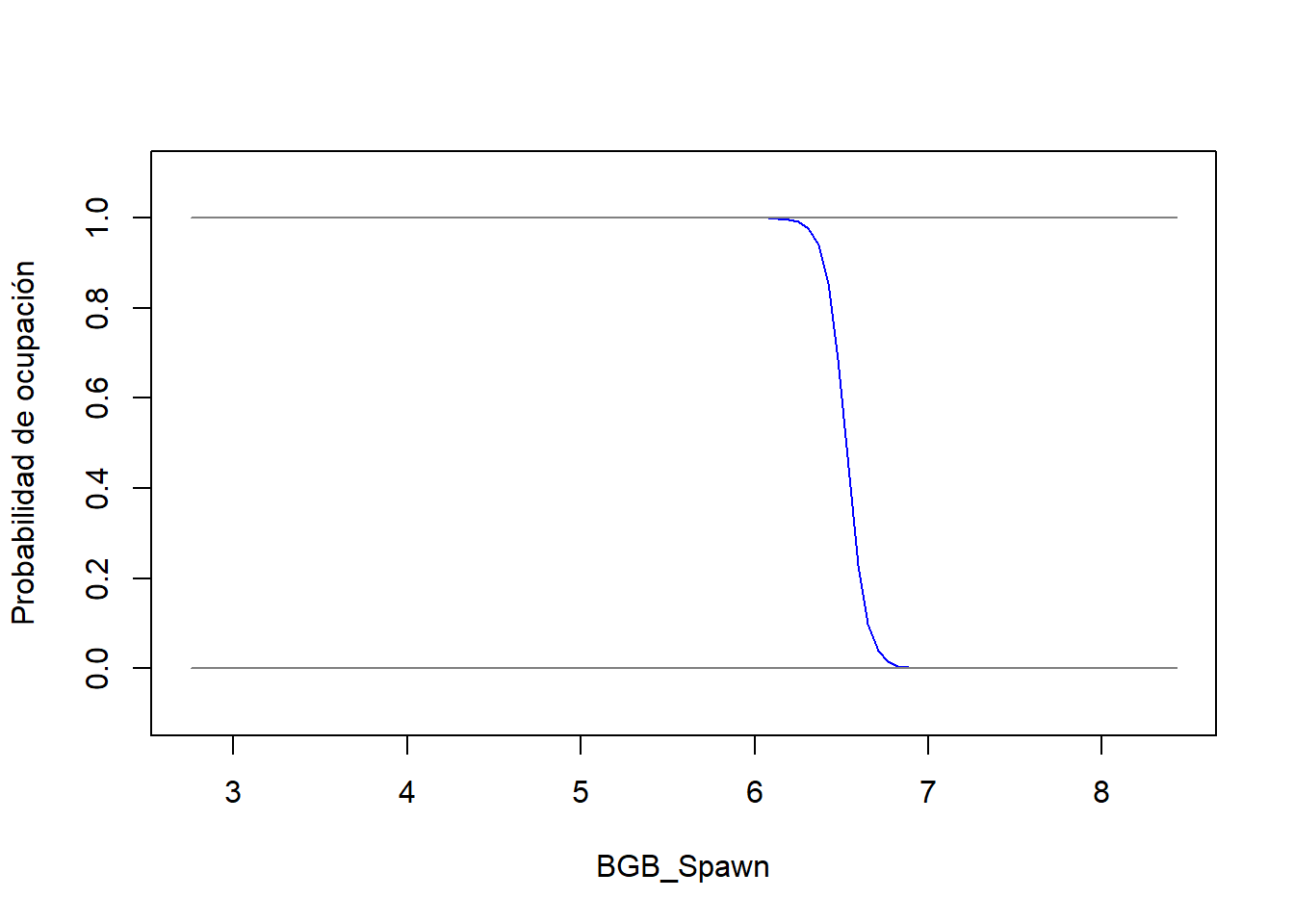
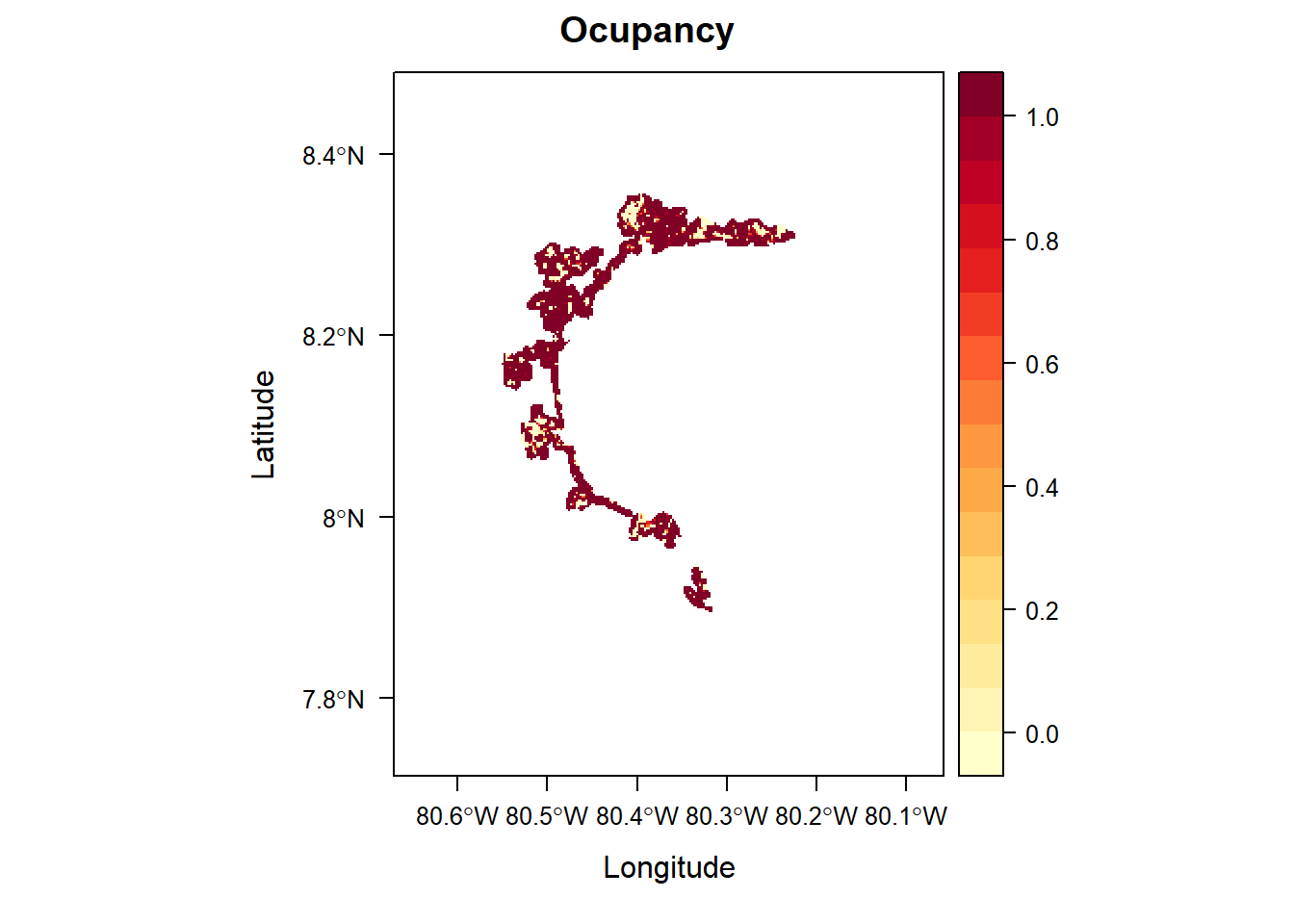
# load function
source("C:/CodigoR/AudubonPanama/R/matrix_creator.R")
############### load Packages
library(readr) # readr
library(readxl) # read excel
library(tidyverse) #
library(unmarked) # occupancy by maximun likelihood
library(sf) # simple feature
library(mapview) # view map
library(ubms) # occupancy by Bayesian made easy
library(stocc) # spatial data for example
library(terra)
library(raster)
library(RColorBrewer)
library(rasterVis)
############### load data
# old
# parita_data_full <- read_delim("C:/CodigoR/AudubonPanama/data/all_records_audubon_panama_to_occu.csv",
# delim = "\t", escape_double = FALSE,
# col_types = cols(eventDate = col_date(format = "%Y-%m-%d")),
# trim_ws = TRUE)
# new
parita_data_full <- read_excel("C:/CodigoR/AudubonPanama/data/all_records_audubon_panama_Darwin_Core_revision.xlsx",
col_types = c("text", "text", "text",
"date", "text", "text", "numeric",
"numeric", "text", "text", "numeric",
"text", "numeric", "numeric", "text",
"numeric", "numeric", "numeric",
"numeric", "numeric", "text", "text",
"numeric", "text", "numeric", "text",
"text", "text", "text", "text", "numeric",
"text", "numeric", "text", "numeric",
"text", "text"))
covs <- read_csv("C:/CodigoR/AudubonPanama/shp/sites_covs_parita_nona.csv")
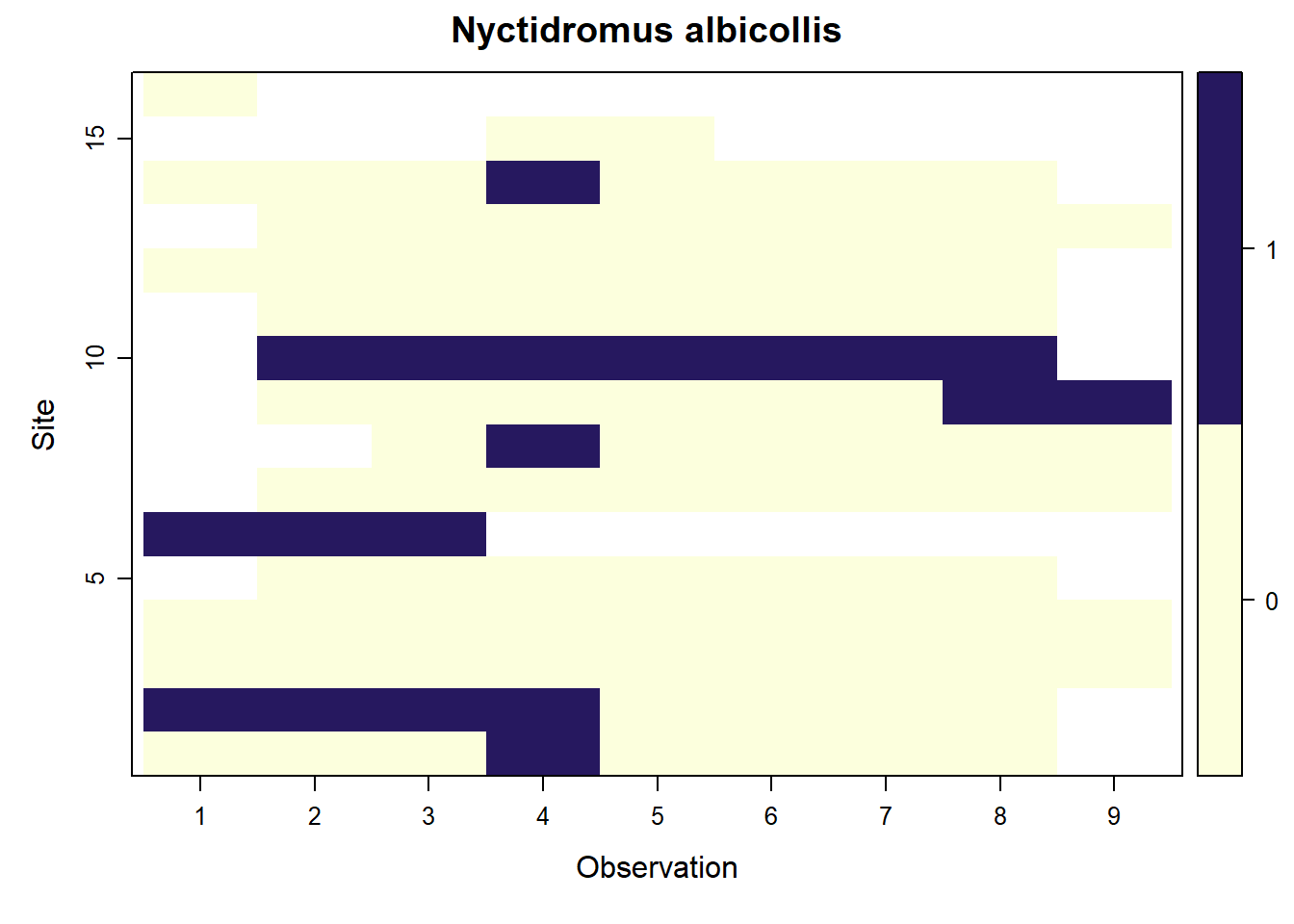
## Nyctidromus albicollis
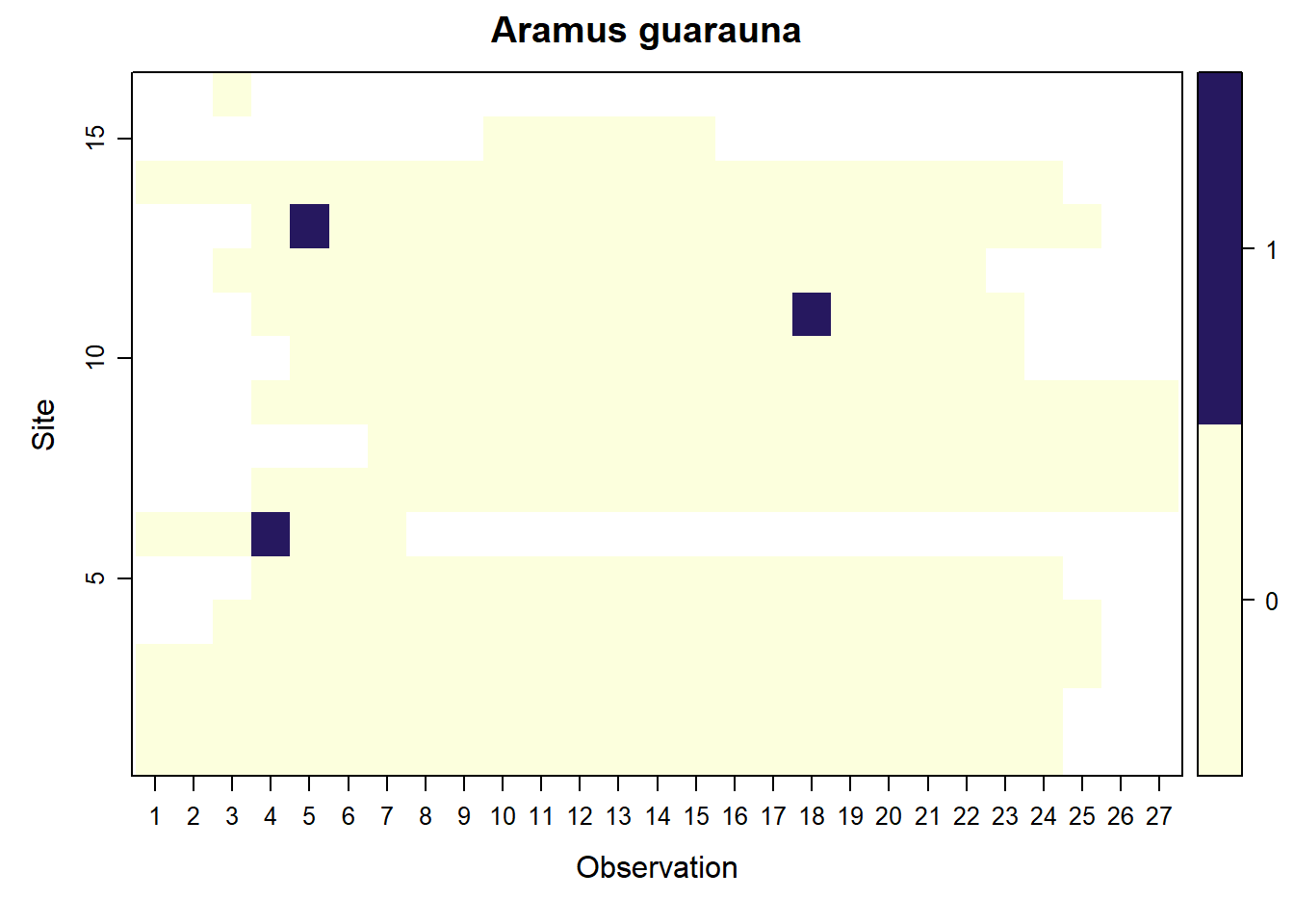
## Aramus guarauna
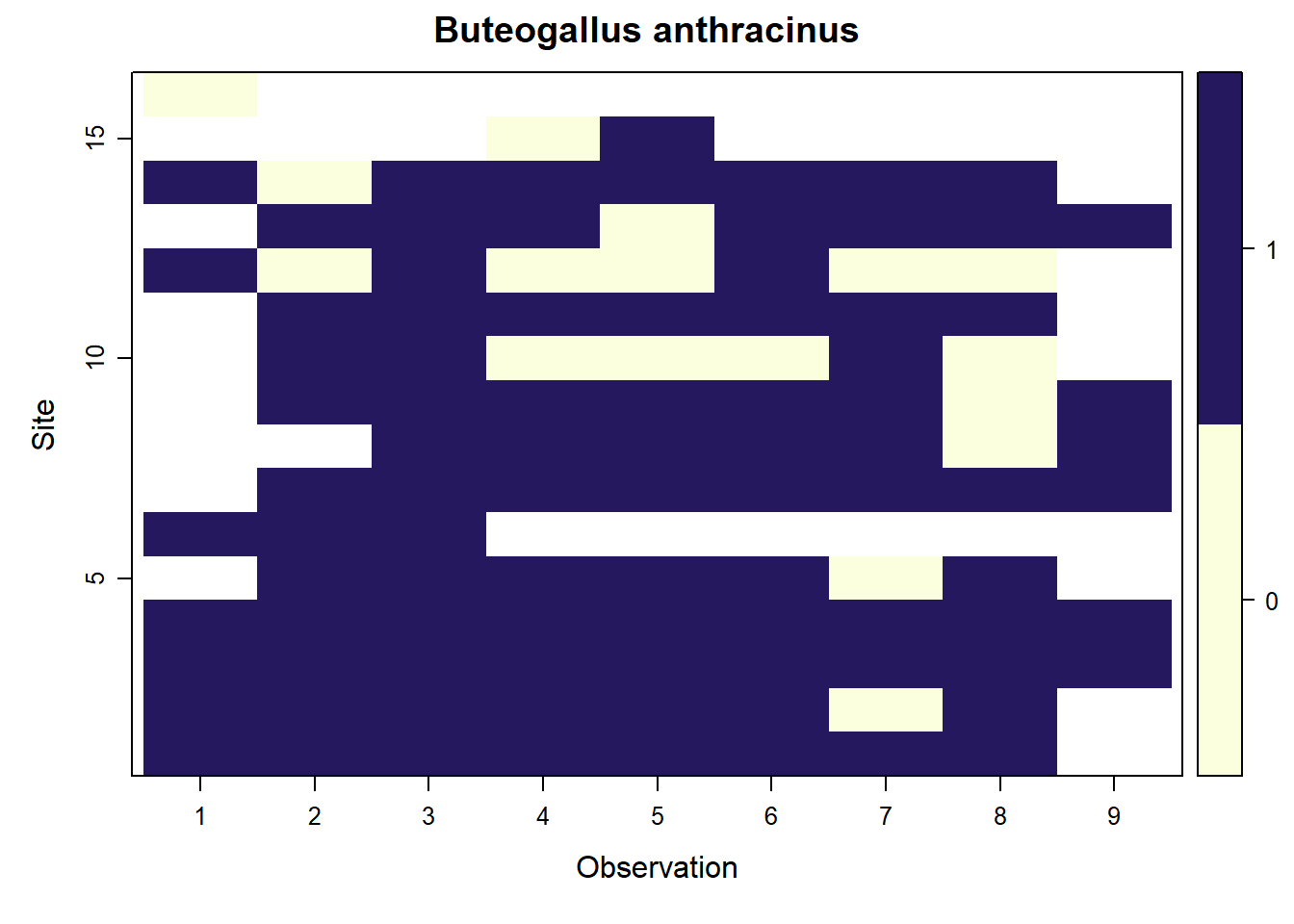
## Buteogallus anthracinus
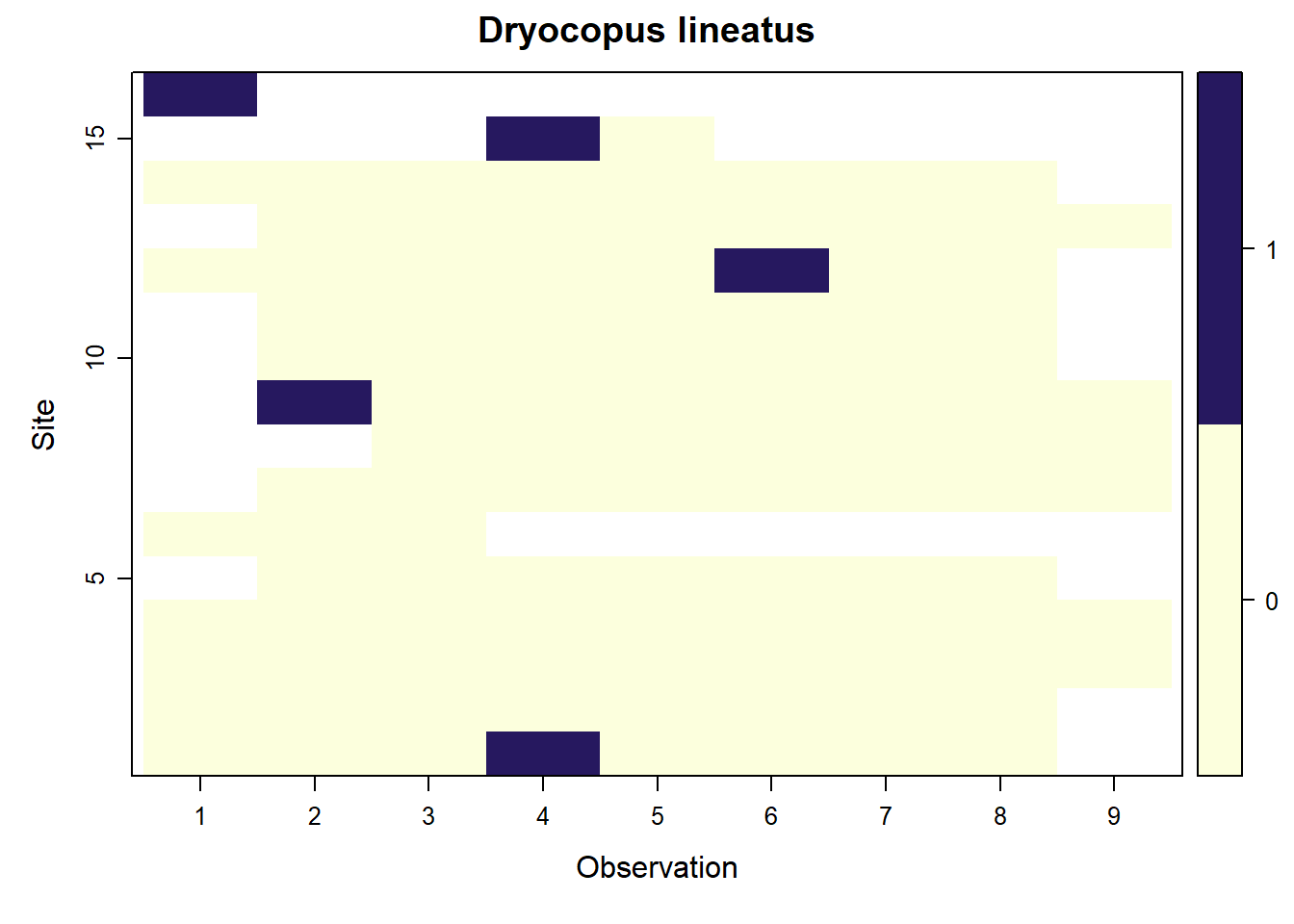
## Dryocopus lineatus
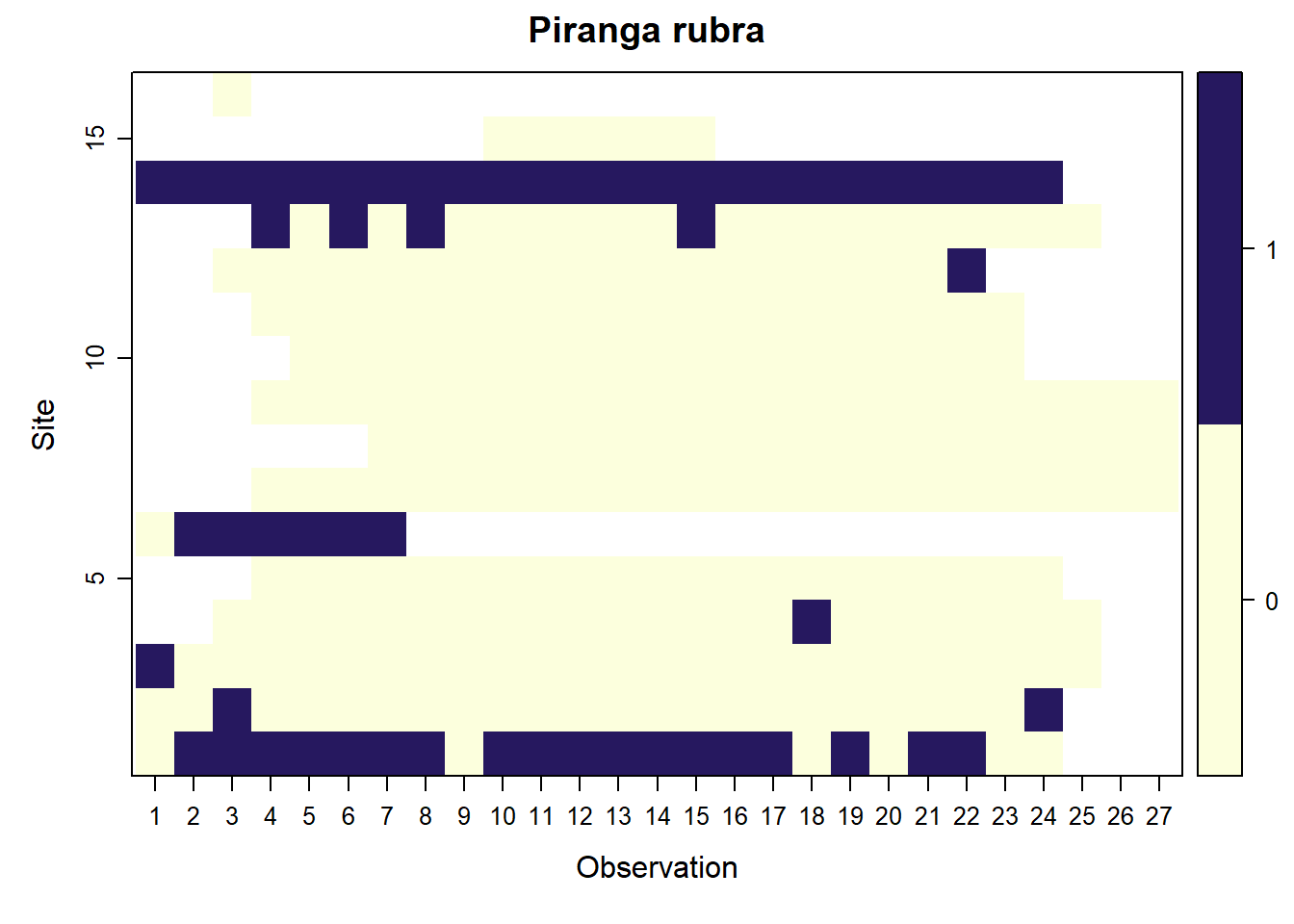
## Piranga rubra
# filter by threshold and species
parita_data_5sp <- parita_data_full %>%
filter(confidence >= threshold) %>%
filter(scientificName == c("Nyctidromus albicollis",
"Aramus guarauna",
"Buteogallus anthracinus",
"Dryocopus lineatus",
"Piranga rubra"
))
# insert Max and min to get matrix limits
# start_date
parita_data2 <- parita_data_5sp %>%
group_by(locationID) %>%
mutate(start_date = min(eventDate),
end_date= max(eventDate))
# apply function to get matrix per species
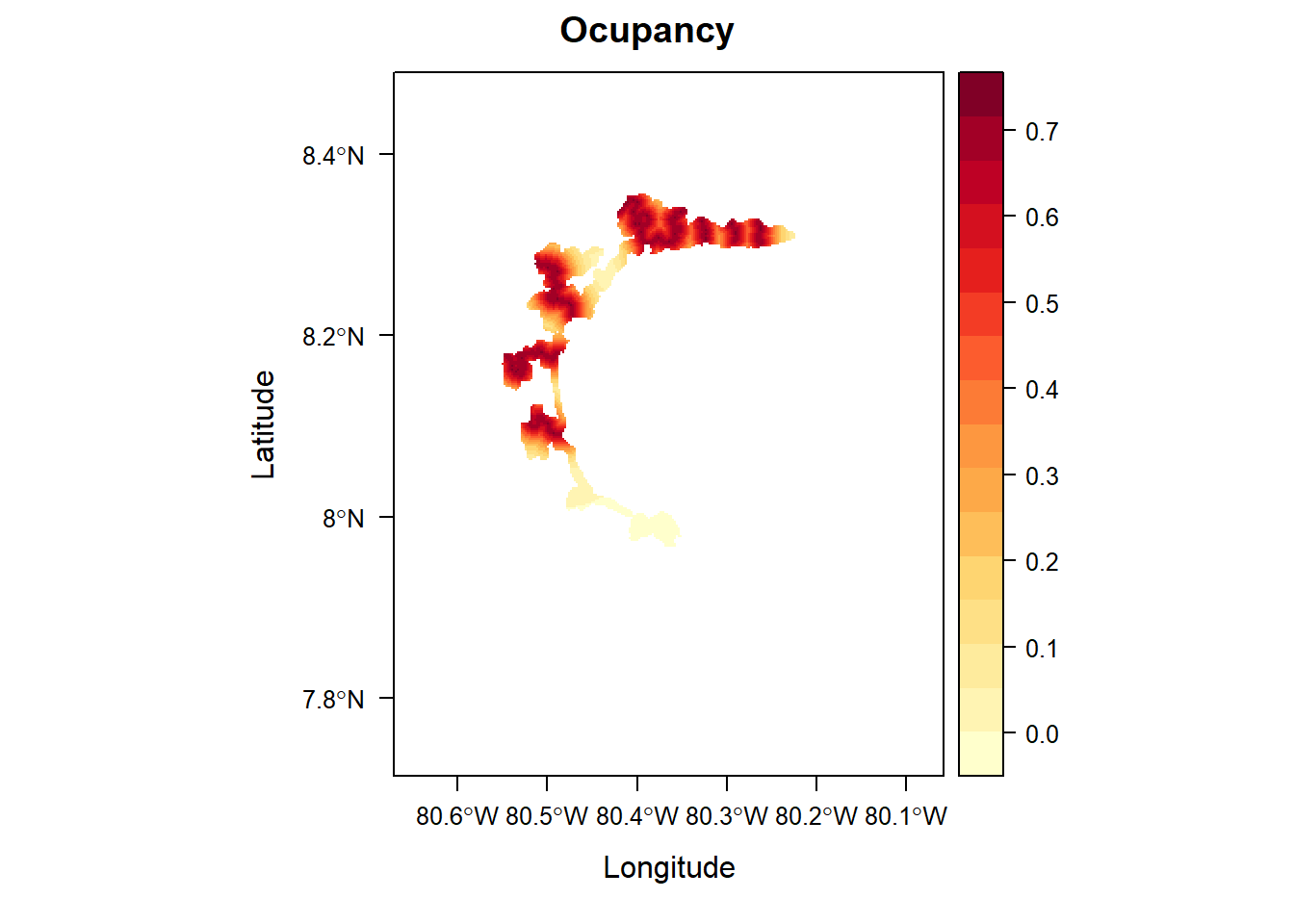
parita <- f.matrix.creator2(data = parita_data2, year = 2023)
# centroide for terra
# centoide <- centroids(puntos, TRUE)
centroide <- c(mean(as.numeric(parita_data_5sp$decimalLongitude)), mean(as.numeric(parita_data_5sp$decimalLatitude)))
clip_window <- extent(-80.67105, -80.05822, 7.713744, 8.489888) # 7.932311, -80.612233 8.360073, -80.180109
bb <- c(-80.612233, -80.180109, 7.932311, 8.360073)
srtm <- raster::getData('SRTM', lon=centroide[1], lat=centroide[2])
# crop the raster using the vector extent
srtm_crop <- raster::crop(srtm, clip_window)
#rast(srtm_crop) # convert to terra
# altitud <- elevation_3s(-72.893262, 7.664081007, path="data")
# altitud <- as.numeric(Camptostoma_obsoletum_occu_dia$Altitude)
# convierte covs a puntos terra
puntos <- vect(covs, geom=c("Longitude", "Latitude"), crs="EPSG:4326")
# convierte a sf
puntos_sf <- sf::st_as_sf(puntos)
# extract elev from raster
cam_covs <- raster::extract(srtm_crop, puntos_sf)
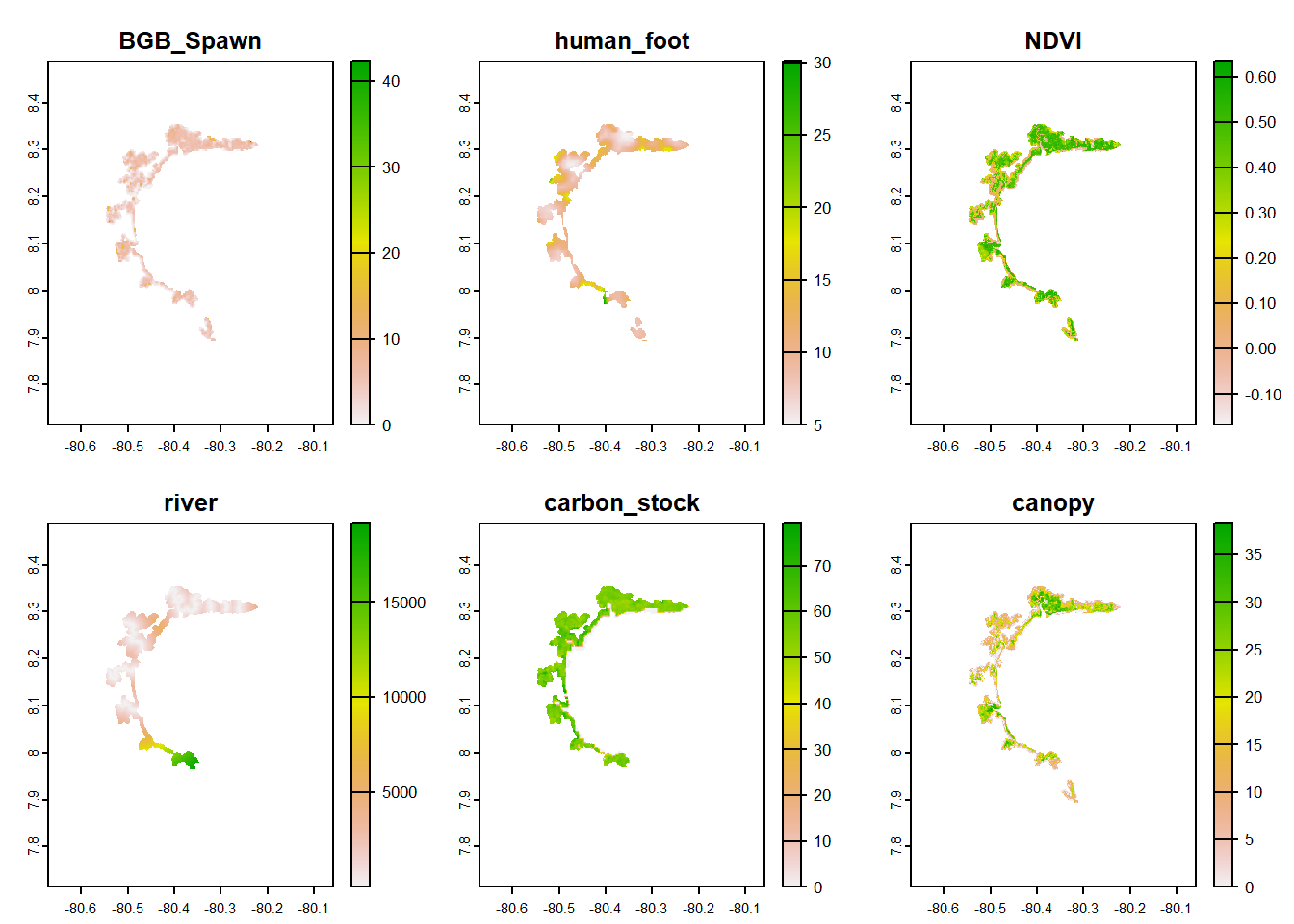
BGB_Spawn <- rast("C:/CodigoR/AudubonPanama/raster/BGB Spawn.tif")
AGB_Spawn <- rast("C:/CodigoR/AudubonPanama/raster/AGB Spawn.tif")
NDVI <- rast("C:/CodigoR/AudubonPanama/raster/S2_NDVI_median_v2.tif")
roads <- rast("C:/CodigoR/AudubonPanama/raster/roads_final_v2.tif")
carbon_stock <- rast("C:/CodigoR/AudubonPanama/raster/soil_organic_carbon_stock_0-30m.tif")
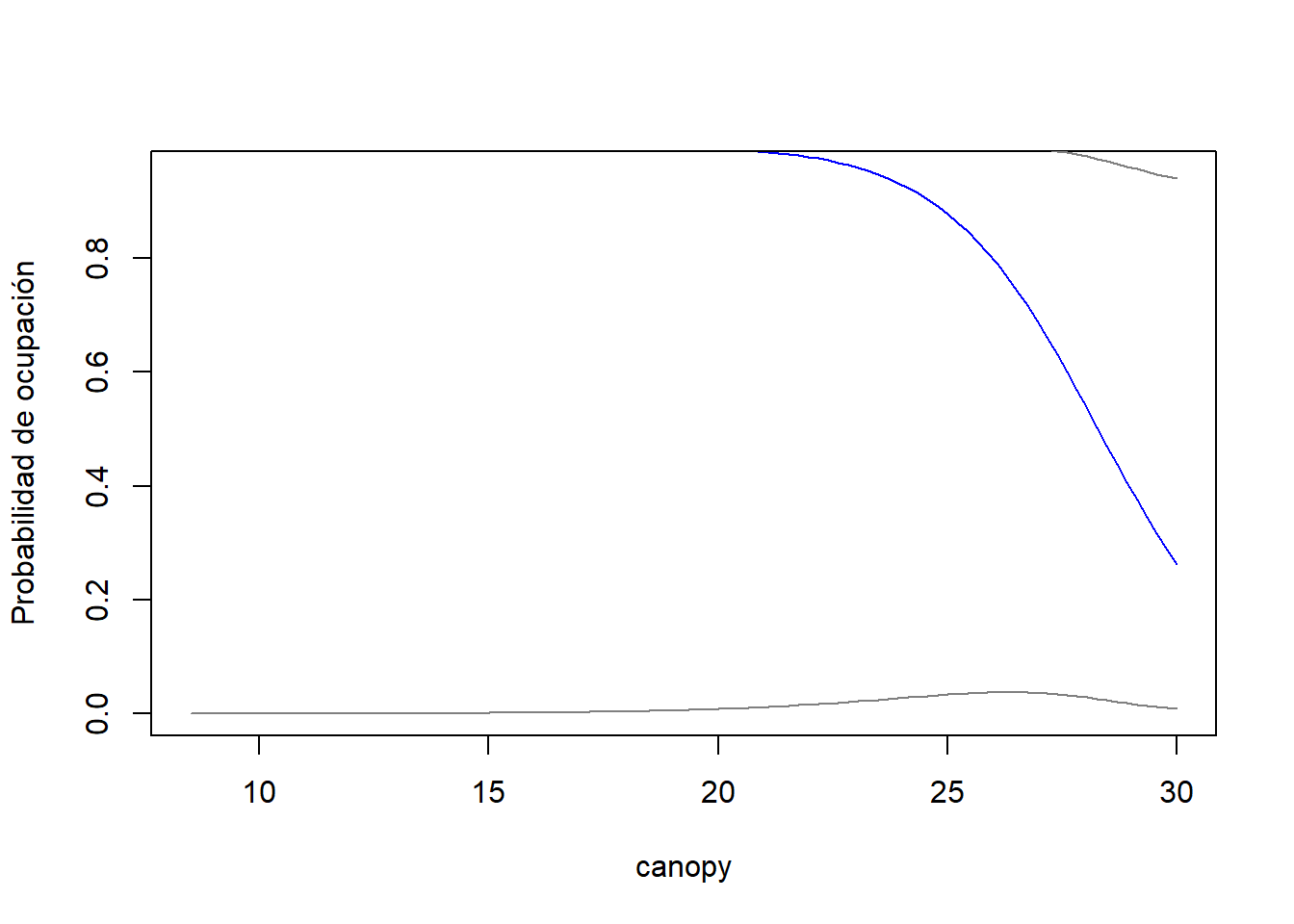
canopy <- rast("C:/CodigoR/AudubonPanama/raster/canopy_height_jetz2.tif")
human_foot <- rast("C:/CodigoR/AudubonPanama/raster/human_footprint.tif")
river <- rast("C:/CodigoR/AudubonPanama/raster/rivers_final_v2.tif")
# make elevation equal
# srtm_projected <- projectRaster(srtm_crop, crs=projection(NDVI), method="ngb")
# elev <- mask(resample(rast(srtm_projected), NDVI), NDVI)
# list of terras SpatRasters
many_rasters <- list(BGB_Spawn, human_foot, NDVI, river, carbon_stock, canopy)
# terra stack
covs_raster <- rast(many_rasters)
names(covs_raster) <- c("BGB_Spawn", "human_foot", "NDVI", "river", "carbon_stock", "canopy")
# change NA to 0
covs_raster0 <- subst(covs_raster, NA, 0)
# extract covariates
covs_raster_val <- extract(covs_raster0, puntos_sf)
plot(covs_raster)